Description
Pintail
Pintail is a free and open-source Java-based app that's super handy for spotting and checking out 16S rRNA chimeras and other kinds of anomalies. It does this by comparing the evolutionary distances between your query and subject sequences, making it a great tool for researchers.
Getting Started with Pintail
When you download Pintail, it comes as an archive without a setup kit. So, all you need to do is extract the files and double-click the JAR executable file to get started. Just remember, you'll also need ClustalW, which is another program used for sequence alignment.
Setting Up ClustalW
You’ll have to specify the full path to ClustalW when using Pintail. It automatically finds the executable file by its name, so if it’s named something else, make sure to rename it to "clustalw.exe" for everything to work smoothly.
User-Friendly Interface
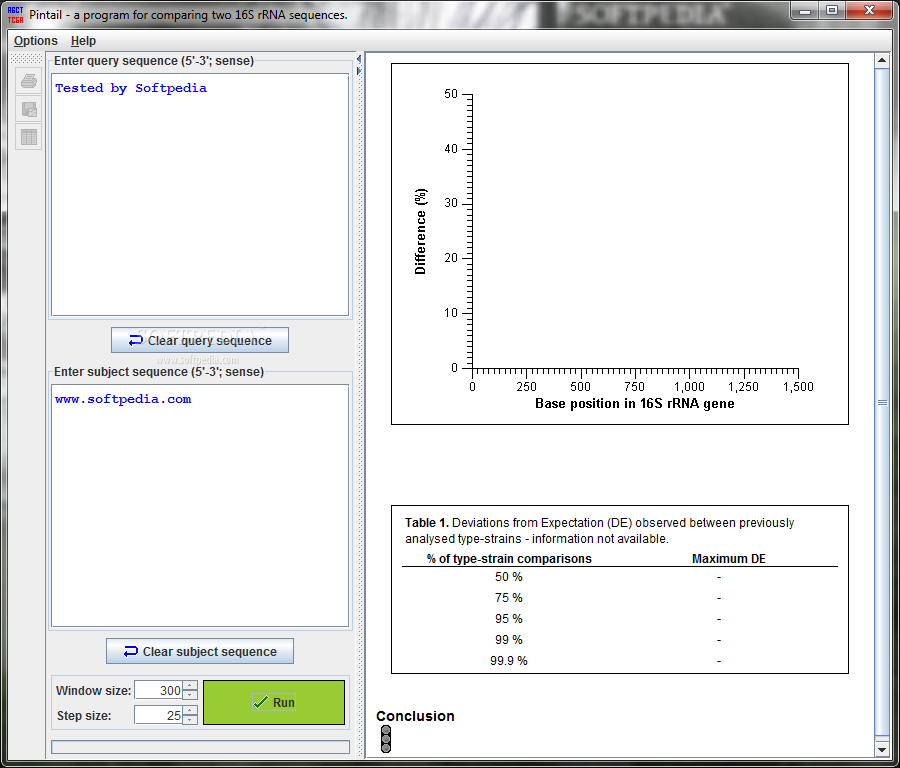
The interface of Pintail is pretty straightforward. You’ll see a single window with text boxes where you can enter your query and subject sequences (5'-3'; sense). Don’t forget that both fields must be filled out before you can generate your plots!
Customizing Your Experience
You can change things like the default window size and step size too! Plus, there are options to view, print, or export your graphs in formats like PostScript, JPEG, or PNG that represent base positions in the 16S rRNA gene.
Understanding Your Results
The information displayed explains what’s happening in your graph. It shows observed versus expected differences along with deviations from expected differences (DE), including details about type-strain comparisons and maximum DEs. This helps you understand any strong evidence of sequence anomalies.
Easy Navigation Tools
Pintail also has handy buttons for undoing and redoing actions along with options for cutting, copying, pasting—basically managing all your data easily! You can choose from three UI themes as well as hide tooltips if they’re not needed. And if you're ever stuck, there's help documentation available that includes tutorials and IUB base codes for reference.
A User-Friendly Experience
Overall, Pintail packs a user-friendly punch! It has approachable features for detecting potential sequence anomalies like 16S rRNA chimeras. In our tests, it ran smoothly and created plots quickly. Just keep in mind that setting up everything might take a bit of time!
User Reviews for Pintail 7
-
for Pintail
Pintail is a powerful tool for identifying and examining sequence anomalies. Its user-friendly interface and swift plots make it a valuable asset.
-
for Pintail
Pintail is an incredible tool for analyzing 16S rRNA sequences. It’s user-friendly and efficient!
-
for Pintail
I love Pintail! The interface is clean, and it provides detailed insights into sequence anomalies.
-
for Pintail
Pintail makes identifying chimeras so easy! The export options are a great feature for my research.
-
for Pintail
Fantastic app! Pintail's ability to visualize data helps in understanding complex sequences effortlessly.
-
for Pintail
Highly recommend Pintail! The setup was simple, and the results are reliable. Great for any researcher!
-
for Pintail
Pintail exceeded my expectations! It’s powerful yet straightforward, perfect for studying 16S rRNA.