Description
ClustalW
ClustalW is a reliable software tool made for genetics pros. It helps you easily handle multiple alignment tasks and even create phylogenetic trees!
Understanding ClustalW
Now, it's worth noting that ClustalW doesn't have a fancy graphical user interface (GUI). This can make it a bit tricky for newbies because you'll need some basic knowledge of using the command line (CMD) to navigate it.
Helpful Documentation
The good news? ClustalW comes packed with extensive help documentation. You can pull it up anytime by entering the right command, which is super useful when you're stuck!
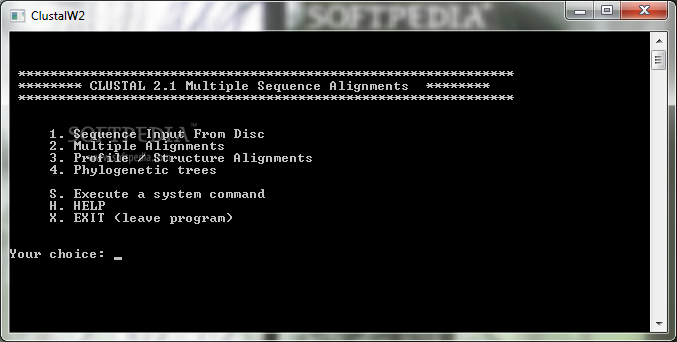
Main Features of ClustalW
This software has several key functions: you can input sequences from your disk, perform multiple alignments, do profile or structure alignments, and even generate phylogenetic trees. Each function is assigned a number from 1 to 4, making it easy to remember what does what.
Supported File Types
ClustalW supports lots of sequence file types! It works with NBRF-PIR, Fasta, ALN (Clustal), Pileup, and GDE files. The cool part is that it often recognizes the file format automatically just by looking at the info at the start or end of your document.
How to Use ClustalW
If you want to add files you want to work on, start by typing ‘1’ and then enter the name of your target item along with its full path. After that, choose your operation by pressing the corresponding number: ‘2’ for aligning sequences or ‘3’ for adding another sequence into an existing one. And if you’re ready to create phylogenetic trees from aligned sequences, just hit ‘4’!
In Summary
To wrap it up, ClustalW is a handy and efficient program that runs in command line mode. It’s here to help you align different DNA sequences into one document without much hassle! If you're ready to give it a try, check out this link for download!
User Reviews for ClustalW 7
-
for ClustalW
ClustalW, though lacking a GUI, offers professionals an effective method for multiple alignments and phylogenetic tree creation. Extensive help docs available.
-
for ClustalW
ClustalW is a game-changer for genetic analysis! The command line interface may seem daunting, but the powerful features and comprehensive documentation make it an invaluable tool for professionals.
-
for ClustalW
Absolutely love ClustalW! It’s efficient and reliable for multiple alignments. The help documentation is thorough, which helps navigate the CMD without much hassle.
-
for ClustalW
ClustalW has transformed my research! Despite the lack of a GUI, its capabilities in handling DNA sequences and generating phylogenetic trees are top-notch. Highly recommend!
-
for ClustalW
This app is fantastic for geneticists! Once you get past the command line interface, it becomes incredibly powerful. The alignment features are spot-on and save so much time!
-
for ClustalW
A must-have tool for anyone in genetics! ClustalW’s ability to align sequences and create phylogenetic trees is impressive. The documentation makes learning easy!
-
for ClustalW
ClustalW is simply amazing! Once you familiarize yourself with the commands, it works like a charm. Great support for various sequence formats too. Five stars!